- Home
- About
- Research
- Education
- News
- Publications
- Guides-new
- Guides
- Introduction to HPC clusters
- UNIX Introduction
- Nova
- HPC Class
- SCSLab
- File Transfers
- Cloud Back-up with Rclone
- Globus Connect
- Sample Job Scripts
- Containers
- Using DDT Parallel Debugger, MAP profiler and Performance Reports
- Using Matlab Parallel Server
- JupyterLab
- JupyterHub
- Using ANSYS RSM
- Nova OnDemand
- Python
- Using Julia
- LAS Machine Learning Container
- Support & Contacts
- Systems & Equipment
- FAQ: Frequently Asked Questions
- Contact Us
- Cluster Access Request
Using Matlab Parallel Server
Introduction
Matlab Parallel Server is a set of Matlab functions that extend the capabilities of the Matlab Parallel Computing toolbox to allow you to submit jobs from your Matlab desktop session directly to the HPC clusters. This allows you to take full advantage of the computing power available on the clusters to solve complex problems while using Matlab on your own computer. In fact, you must use the Matlab Parallel Server if you want to run jobs that span multiple nodes.
When you submit a Matlab Parallel Server job from your Matlab desktop, scripts and data are transferred to the cluster by Matlab. Results from jobs that run on the cluster are readily available to Matlab running on your desktop.
While it is possible to submit a Matlab Parallel Server job as a standalone sbatch script from a cluster head node, the process is somewhat complicated, so we won't be digging into that here. The instructions on this page are for people who want to interact with Parallel Server from their own desktop. Instructions for submitting Matlab Parallel Server jobs using sbatch scripts on the headnode are not yet ready.
- Prerequisites
- Download and Unpack the Cluster Plugin Scripts
- Run the configCluster.m Script to Create a Cluster Profile
- Verify that configCluster.m Created a Cluster Profile
- Working with Cluster Objects
- Examples:
Example 1: A Serial Job
Example 2: A Parallel Job on 1 Node
Example 3: A Distributed Parallel Job on Multiple Nodes
Prerequisites
- Your Matlab version must be R2017a or later.
- You must have the Matlab Parallel Computing Toolbox installed. To see what toolboxes are installed, type the ver command in the Matlab command window. You should see Parallel Computing Toolbox listed.
- You must be connected to the ISU VPN in order to submit jobs from your desktop, regardless of whether you are on-campus or off-campus. (This is because the Matlab cluster integration tools do not support multi-factor authentication such as Okta. The VPN connection helps enforce multi-factor authentication.)
- You must ssh to the cluster at least once before trying to submit any jobs from Matlab.
Download the Cluster Plugin Scripts
In order to submit jobs from your desktop, you will need to install some custom Matlab plugin scripts that know how to interact with the Slurm job scheduler on a cluster. There is a separate cluster plugin script bundle for each cluster (Nova, Condo, and HPC-class). Each bundle contains configuration files and scripts packaged together as .zip files (for Windows and Mac) and as .tar.gz files (for Linux). Use the links below to download the appropriate script bundle you need based on the cluster(s) you plan to use from the links below.
HPC-class:
hpc-class-config-cluster.zip (for Windows and Mac)
hpc-class-config-cluster.tar.gz (for Linux)
Condo:
condo-config-cluster.zip (for Windows and Mac)
condo-config-cluster.tar.gz (for Linux)
Nova:
nova-config-cluster.zip (for Windows and Mac)
nova-config-cluster.tar.gz (for Linux)
Unpack the Cluster Plugin Bundle(s) to a Path on Your Computer
Copy the integration bundle file to a new folder on your computer, such as Downloads/matlab-parallel-server. If you need more than one configuration bundle (i.e. you have access to multiple clusters), you should put the bundle for each cluster in a separate folder (e.g. Downloads/matlab-parallel-server/condo, Downloads/matlab-parallel-server/nova, etc.). Unpack each plugin bundle file into the path you choose.
Run the configCluster.m Script to Create a Cluster Profile
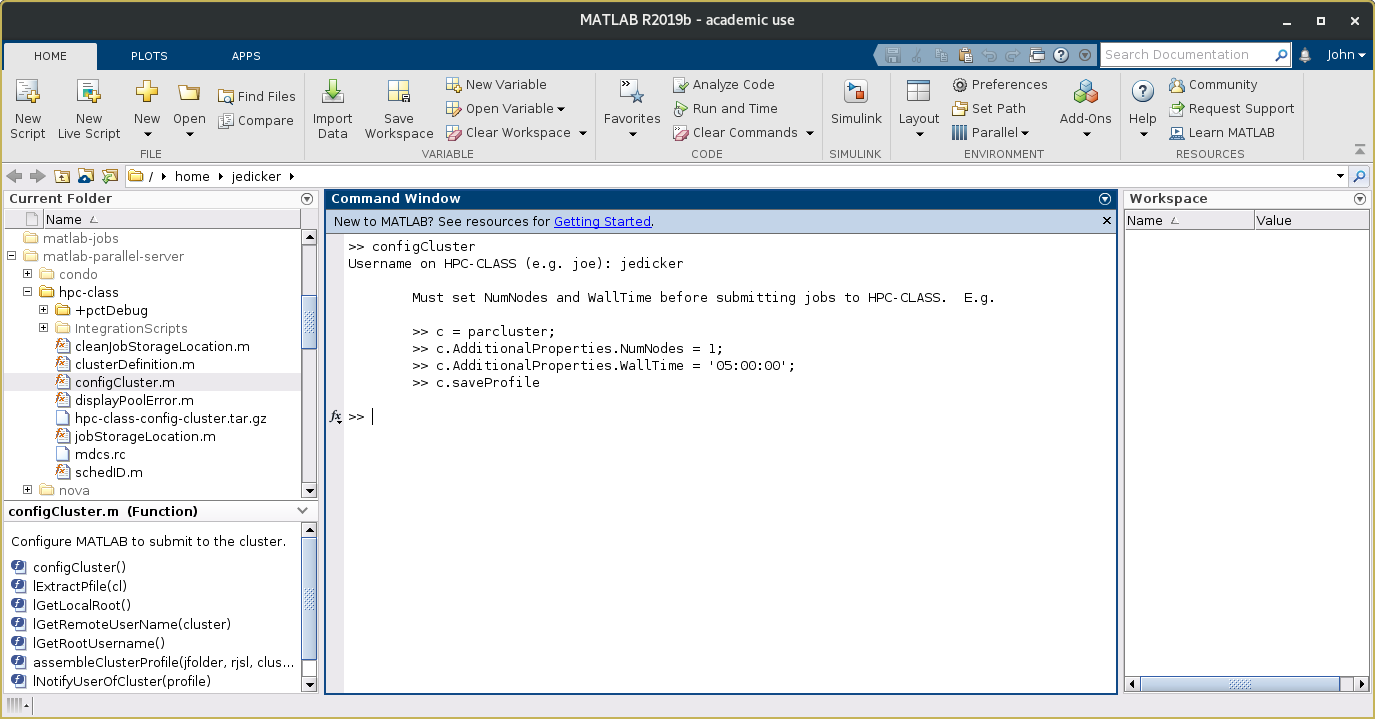
Open Matlab. In the Current Folder window, navigate to the path where you unpacked one of the cluster plugin bundles. In that path, find the Matlab script called configCluster.m. Run this script by right-clicking on the name and selecting Run. (If Matlab asks you if you want to add the path to your environment, select Add Path.)
The configCluster script will prompt you to enter the username you use to log in to the cluster. Just enter your ISU username (NetID). Don't append "@iastate.edu" to it. After you enter your username, the program will display an informational reminder that you will need to set the NumNodes and WallTime parameters before you can submit a job. We'll cover how to set job parameters in the next section. A screenshot of a successful run of the HPC-class version of configCluster is shown below:
Verify that clusterConfig Created a Cluster Profile
If the configCluster program worked correctly, you should see the cluster profile added to your Parallel environment. To see this, select the Home tab in the Matlab desktop. Find the ENVIRONMENT pane. Click on the Parallel pull-down menu and select Create and Manage Clusters. This will open the Cluster Profile Manager window.
Note that the clusterConfig script automatically appends the version of Matlab you are using to the profile name. If you run multiple versions of Matlab, or upgrade to another version, you will need to run the configCluster program again for each Matlab version you use. The Cluster Profile Manager window should look similar to the screenshot below.
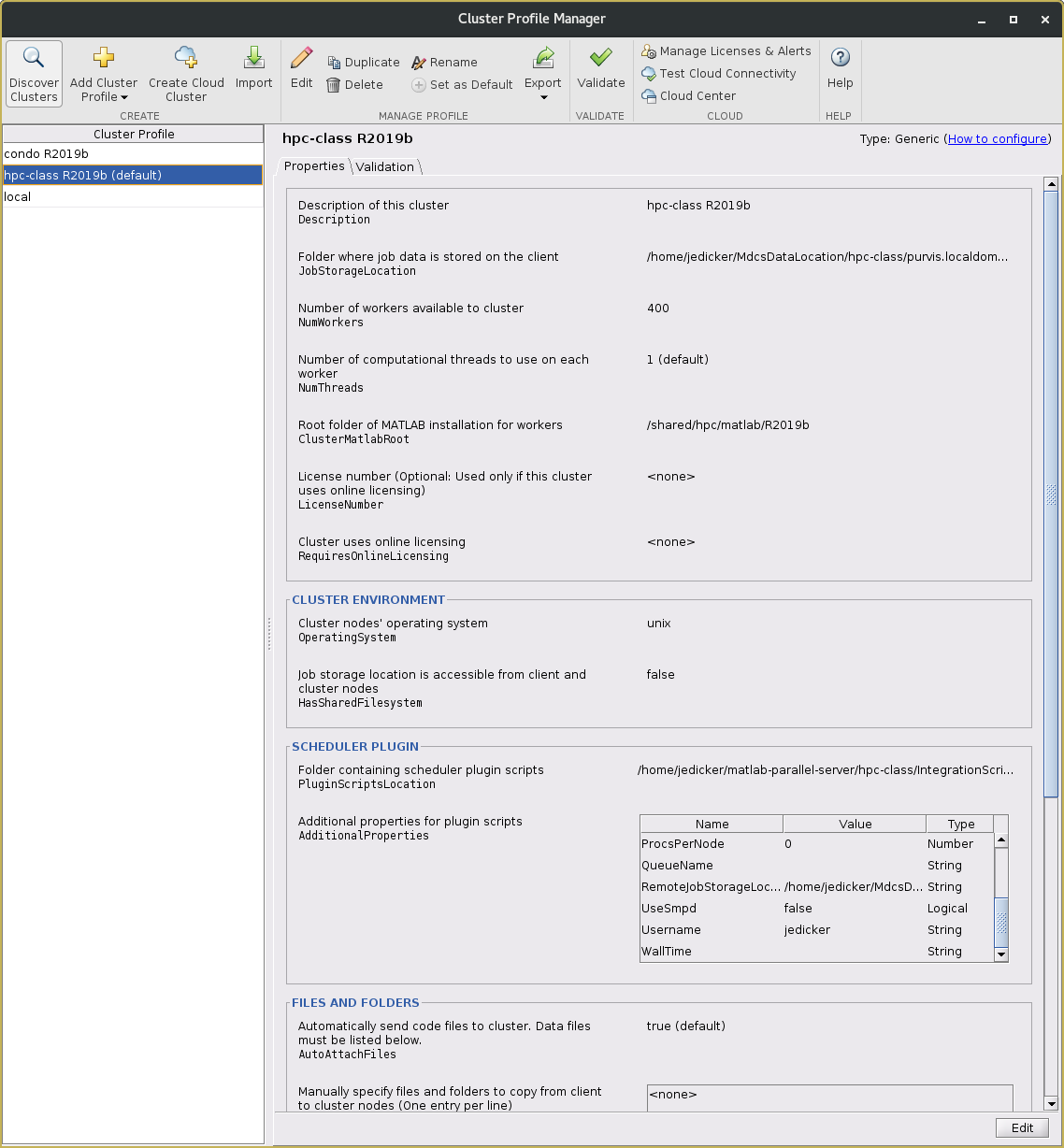
Notice that in the example above 'hpc-class 2019b' is the default cluster configuration. If you have configured multiple cluster profiles, you can specify which one is the default by right-clicking on the desired cluster profile and selecting Set as Default.
Working with Cluster Objects
Using cluster objects is central to using Matlab Parallel Server. The parcluster command is used to initialize a cluster object for your session:
>> c = parcluster('hpc-class R2019b');
>> c
Generic Cluster
Properties:
Profile: hpc-class R2019b
Modified: false
Host: localhost
NumWorkers: 400
NumThreads: 1JobStorageLocation: /home/jedicker/MdcsDataLocation/hpc-class/purvis.localdomain/R2019b/remote
ClusterMatlabRoot: /shared/hpc/matlab/R2019b
OperatingSystem: unixRequiresOnlineLicensing: false
PluginScriptsLocation: /home/jedicker/matlab-parallel-server/hpc-class/IntegrationScripts/h...
AdditionalProperties: List propertiesAssociated Jobs:
Number Pending: 0
Number Queued: 0
Number Running: 0
Number Finished: 8>>
Above, we first created a cluster object 'c' using the 'hpc-class R2019b' profile. Then, we entered 'c' without a semi-colon to see the list of properties in the cluster object. Notice that there is a very important subset of properties called AdditionalProperties. You will need to modify some of these properties values to actually submit a job to Slurm. Let's examine the AdditionalProperties:
>> c.AdditionalProperties
ans =
AdditionalProperties with properties:
AdditionalSubmitArgs: ''
ClusterHost: 'hpc-class.its.iastate.edu'
EmailAddress: ''
EnableDebug: 0
MemUsage: ''
NumNodes: 0
ProcsPerNode: 0
QueueName: ''
RemoteJobStorageLocation: '/home/jedicker/MdcsDataLocation/hpc-class/purvis.localdomain/R2019b/remote'
UseIdentityFile: 0
UseSmpd: 0
Username: 'jedicker'
WallTime: ''
As you can see, when you first create the cluster object, many of the properties do not have values. As a minimum, you must set values for NumNodes, ProcsPerNode, and WallTime. Optionally, you can set the EmailAddress, QueueName, and AdditionalSubmitArgs. The AdditionalSubmitArgs is used for passing any additional options to the Slurm sbatch command.
Examples
The examples below show how to use Matlab Parallel Server to run serial (single CPU) jobs, multi-processor jobs on a single machine, and distributed parallel jobs running on multiple nodes. If you'd like to download Matlab scripts used in the examples, they are available here:
Example 1: Running a Simple Serial Job
In the first example, we will run a simple Matlab job that uses a single processor. This program isn't parallelized, but we can still execute the job on the cluster. We'll begin by creating a Matlab script called mywave.m simply calculates a million points on a sine wave in a for loop. The file mywave.m has been created in our current Matlab folder. The function is pretty simple:
%-- mywave.m --%
% a simple 'for' loop (non-parallelized):
for i = 1:1000000
A(i) = sin(i*2*pi/102400);
end
As it is, this function just loops a million times. To run this job on the cluster, we'll create a new script called 'run_serial_job.m':
% run_serial_job
% Run a serial (i.e. single CPU, non-parallelized) Matlab job on a cluster.
% Calls a script 'mywave' that executes a loop in serial.
% First initialize a cluster object based on an existing cluster profile.
c = parcluster('condo R2019b')
% Use the AdditionalProperties property of the cluster object to set job specific details:
c.AdditionalProperties.NumNodes = 1; % Number of nodes requested
c.AdditionalProperties.EmailAddress = 'your_netid@iastate.edu'; % Your Email address (please modify accordingly).
c.AdditionalProperties.ProcsPerNode = 1; % Number of processors per node.
c.AdditionalProperties.WallTime = '2:00:00'; % Max wall time
c.AdditionalProperties.QueueName = 'compute';
c.AdditionalProperties.AdditionalSubmitArgs = '';
% Other properties you may need to set:
% To set a specific queue name:
% c.AdditionalProperties.QueueName = 'freecompute';
% To set the Slurm job name:
% c.AdditionalProperties.AdditionalSubmitArgs = '--job-name=xxx';
% NOTE: if --job-name is not set here then Matlab assigns the job name itself as "JobN" where
% N is determined by Matlab.% Before starting the job, start a job timer to see how long the job runs:
tic
% Below, submit a batch job that calls the 'mywave.m' script.
% Also set the parameter AutoAddClientPath to false so that Matlab won't complain when paths on
% your desktop don't exist on the cluster compute nodes (this is expected and can be ignored).myjob = batch(c,'mywave','AutoAddClientPath',false)
% see https://www.mathworks.com/help/parallel-computing/batch.html for additional tips and examples for using the batch command.
% Wait for the job to finish (on a busy server, this might not be a good strategy).
wait(myjob)
% display the job diary (i.e. the Matlab standard output text, if any)
diary(myjob)
% load the 'A' array (computed in mywave) from the results of job 'myjob':
load(myjob,'A');
%-- plot the results --%
plot(A);
% print the elapsed time for the job:
toc
When you run this script in Matlab, it should prompt you for your password for logging in to the cluster. You will not need your google authenticator verification code (since you should already be connected over from the VPN). Matlab caches your password during this session so you don't have to enter your again for submitting other jobs during the same Matlab session.
Example 2: Parallel job on a single node
In the next example, we will run a parallel job using 8 processors on a single node. We will be using a parallelized version of mywave.m called parallel_mywave.m that uses the parfor statement to parallelize the previous for loop:
%-- parallel_mywave --%
% A parfor loop will use parallel workers if available.
parfor i = 1:10000000
A(i) = sin(i*2*pi/2500000);
end
The parfor command will use a pool of workers to execute the loop in parallel if a worker pool exists. You can learn more about the use of parfor loops here: https://www.mathworks.com/help/parallel-computing/parfor.html
Now we'll use a new Matlab script run_parallel_job.m (shown below) to run this job. Note that the batch command has changed significantly by adding a pool for 8 workers. Consequently, the AdditionalProperties.ProcsPerNode property is set to 1 more than the number of workers:
% run_parallel_job
% Run a parallel Matlab job on a 1 cluster node with a pool of 8 Matlab workers.% First initialize a cluster object based on an existing cluster profile,
% in this case we are using the 'condo R2019b' profile.
c = parcluster('condo R2019b')
% Use the AdditionalProperties property of the cluster object to set job specific details:
c.AdditionalProperties.NumNodes = 1; % Number of nodes requested.
c.AdditionalProperties.EmailAddress = 'your_netid@iastate.edu'; % Your Email address (please modify).
c.AdditionalProperties.ProcsPerNode = 9; % 1 more than number of Matlab workers per node.
c.AdditionalProperties.WallTime = '2:00:00'; % The max wall time for the job.
c.AdditionalProperties.QueueName = 'compute';
c.AdditionalProperties.AdditionalSubmitArgs = '';% Examples of other properties that you might need to set:
% To set a specific queue name, in this case to use the 'freecompute' free tier:
% c.AdditionalProperties.QueueName = 'freecompute';
%
% To set the Slurm job name. (if not set, Matlab will use "JobN" where N is determined by Matlab):
% c.AdditionalProperties.AdditionalSubmitArgs = '--job-name=xxx';
% NOTE: The value of AdditionalProperties.AdditionalSubmitArgs is simply added on to the sbatch command
% so this can be used supply any additional options to sbatch.
% Start a job timer for recording the elapsed time for the job:
tic
% The batch command below creates a job object called 'myjob' that runs a
% Matlab job with 8 parallel pool workers.
% NOTE: Matlab will add an additional worker to the pool for its own use so be
% sure that the number of processors requested from Slurm (NumNodes X ProcsPerNode)
% is greater than the total number of workers needed by Matlab.
% We also set the parameter AutoAddClientPath to false so that Matlab won't complain when paths on
% your desktop don't exist on the compute node (this is typical and can be ignored).
myjob = batch(c,'parallel_mywave','pool', 8, 'AutoAddClientPath',false)
% see https://www.mathworks.com/help/parallel-computing/batch.html for additional tips and examples.
% Wait for the job to finish before continuing.
wait(myjob)
% load the 'A' array from the job results. (The values for 'A' are calculated in parallel_mywave.m):
load(myjob,'A');
%-- plot the results --%
plot(A);% print the elapsed time for the job:
toc
Example 3: Distributed parallel job on multiple nodes
The next example, run_distributed_parallel_jobs.m, executes a function in parallel using 48 workers distributed to 4 nodes and 12 cpus per node.
% run_distributed_parallel_job.m
% Run a parallel Matlab job on a cluster with a pool of 48 Matlab workers across 4 compute nodes.% First initialize a cluster object based on an existing cluster profile,
% in this case we are using the 'condo R2019b' profile.
c = parcluster('condo R2019b')% Use the AdditionalProperties property of the cluster object to set job specific details:
c.AdditionalProperties.NumNodes = 4; % Number of nodes requested
c.AdditionalProperties.EmailAddress = 'your_netid@iastate.edu'; % Your Email address (please modify).
c.AdditionalProperties.ProcsPerNode = 13; % Number of processors per node (1 more than Matlab workers per node).
c.AdditionalProperties.WallTime = '3:00:00'; % Set a maximum wall time of 3 hours for this job.
c.AdditionalProperties.QueueName = 'compute';
c.AdditionalProperties.AdditionalSubmitArgs = '';% Other properties often needed:
% To set a specific queue name, in this case to use the 'freecompute' free tier:
% c.AdditionalProperties.QueueName = 'freecompute';
% To set the Slurm job name. (if not set, Matlab will use "JobN" where N is determined by Matlab):
% c.AdditionalProperties.AdditionalSubmitArgs = '--job-name=xxx';
% NOTE: The value of AdditionalProperties.AdditionalSubmitArgs is simply added on to the sbatch command.% Start a job timer for recording the elapsed time for the job:
tic% The batch command below creates a Matlab job with 48 pool workers, one
% per CPU. Each worker will do a portion of the computation in the parallel_eigen function.
% A handle to the job object is created called 'myjob'.
% NOTE: Matlab will add an additional worker to the pool for its own use so be
% sure that the number of processors requested from Slurm (NumNodes X ProcsPerNode)
% is at least 1 greater than the total number of workers needed by Matlab.
% Also, set the parameter AutoAddClientPath to false so that Matlab won't complain when paths on
% your desktop don't exist on the compute node (this is typical and can be ignored).myjob = batch(c,'parallel_eigen','pool', 48, 'AutoAddClientPath',false)
% see https://www.mathworks.com/help/parallel-computing/batch.html for additional tips and examples.
% Wait for the job to finish (on a busy cluster, this may not be a good strategy).
wait(myjob)
% load the 'E' array from the job results. (The values for 'E' are calculated in parallel_eigen.m):
load(myjob,'E');
%-- plot the results --%
plot(E);
% print the elapsed time for the job:
toc